Institute of Applied and Computational Mathematics
IACM is one of the few research institutes in Europe dedicated to promoting the use of advanced mathematics in natural sciences and engineering
Institute of Applied and Computational Mathematics
IACM is one of the few research institutes in Europe dedicated to promoting the use of advanced mathematics in natural sciences and engineering
Data Science
ABOUT
RESEARCH AND DEVELOPMENT ACTIVITIES
-
Deep Learning and Generative AI
Deep learning: Deep learning has recently been at the forefront of artificial intelligence (AI) research and has truly revolutionized several fields of AI. Data Science Group members are actively involved in developing methods that can help us both better understand the limitations of existing deep learning approaches and also improve various aspects of their performance. They have worked on a wide variety of topics from this area, exploring both fundamental questions and practical applications. This includes, to mention a few of these topics, exploring and proposing novel deep network architectures, developing new ways of effectively transferring knowledge between networks, revisiting weight parameterizations for deep networks in order to improve their generalization capabilities, proposing novel self-supervised and few-shot learning methods, devising and applying learning approaches that advance the state-of-the-art for fundamental problems from the areas of computer vision and image analysis, making use of attention in the context of knowledge distillation, proposing hybrid (scattering-based) convolutional network architectures that allow for better representation learning and more interpretable features, as well as properly adapting deep neural networks such that they can be applied to arbitrary graph-structured data directly and can also handle structured-prediction tasks.
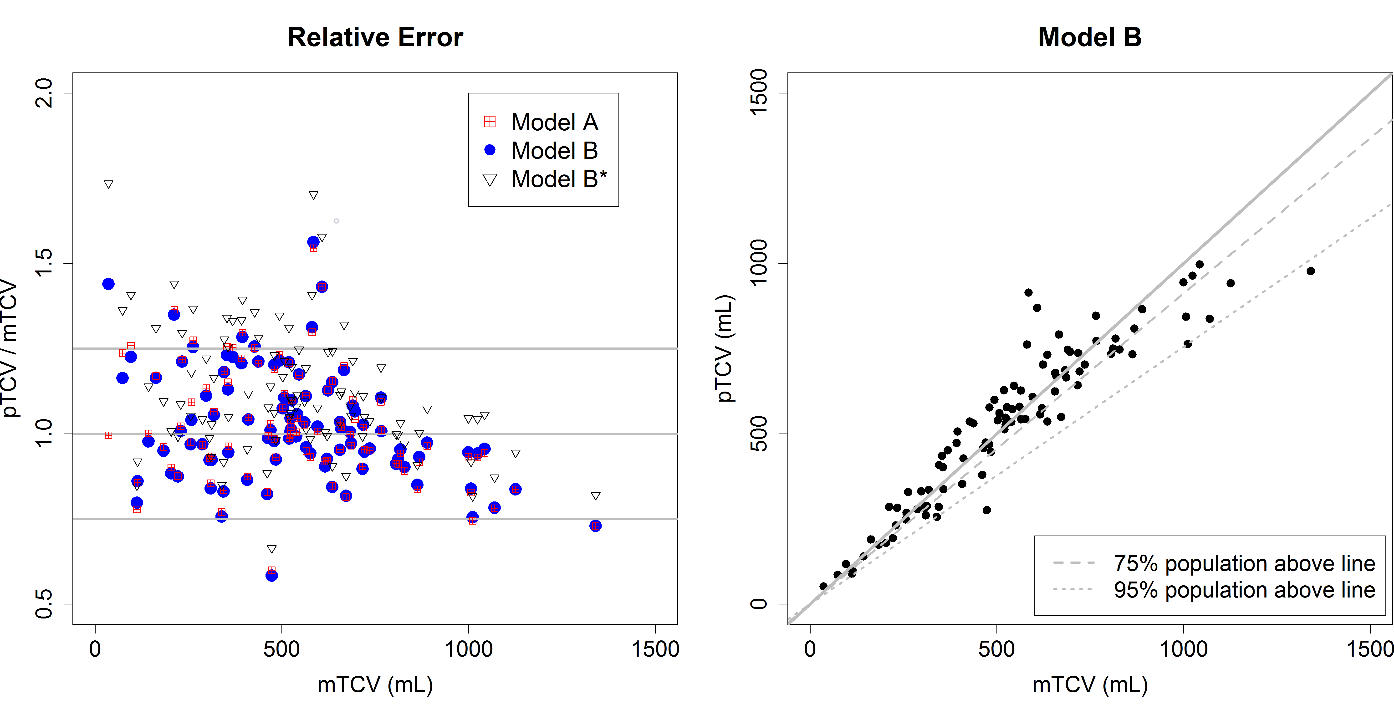
Neural-based Speech Synthesis - Deep Learning in Speech Processing: Deep Neural Networks have taken the engineering community by storm. Data-rich areas such as image processing and speech processing have been transformed during the last years. The Data Science Group combines its expertise on speech processing and applies deep learning techniques to applications such as voice conversion, speech synthesis and speech enhancement.
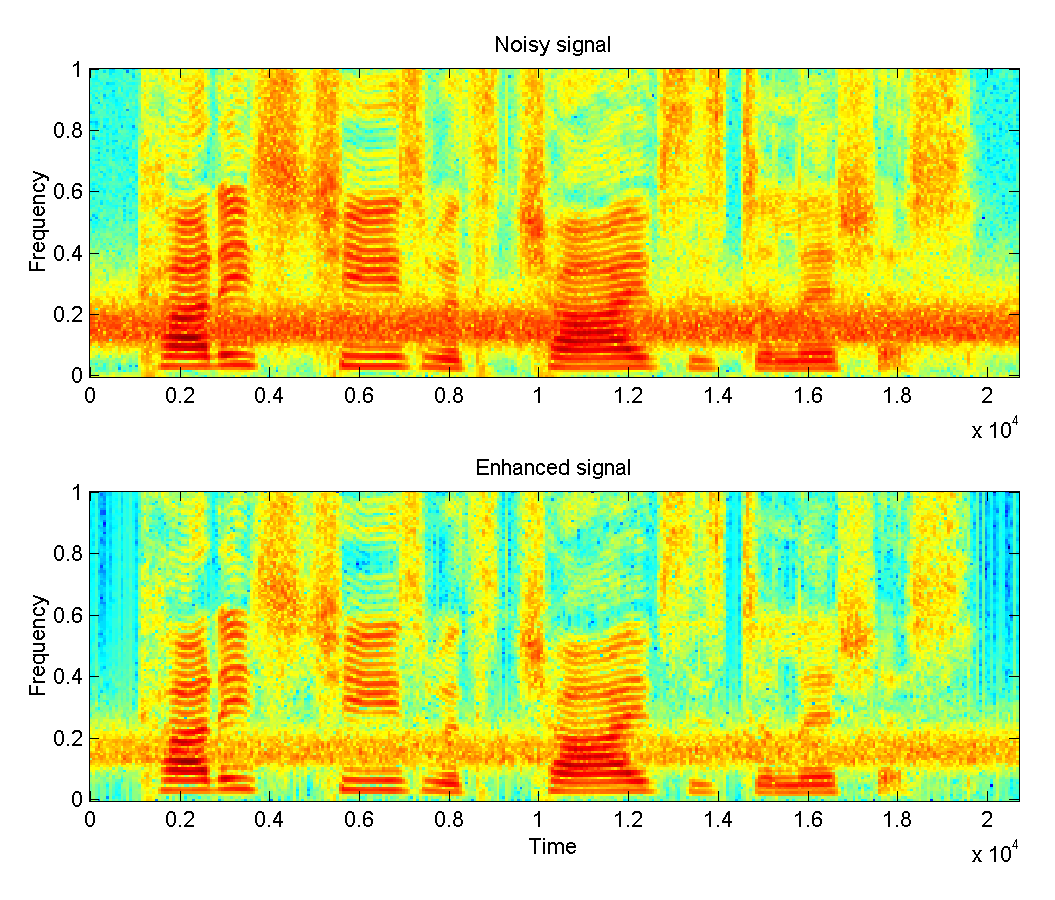
Generative Adversarial Nets - Deep Generative Learning: Generative models based on Deep Neural Nets have shown unprecedented capabilities in sampling data from complex but unknown distributions. Researchers in the Data Science Group develop novel algorithms for training generative models, focusing on Generative Adversarial Networks (GANs). GANs have been used in data augmentation schemes to generate synthetic data that follow the same distributional characteristics as the original dataset (which may contain sensitive information or limited number of cases). The proposed methodology has been applied to identify dyslexia in children, using measurements from specialized eye trackers.
- Spatial, Temporal & Spatio-temporal Statistics
- High-Dimensional and Sparse Statistics
- Uncertainty Quantification
- Additional Topics
- Journal Club on AI
- Education and Training
Data Science
RESEARCH AND DEVELOPMENT PROGRAMS
A. ONGOING PROJECTS
- Title: TURING: Trustworthy Unified Robust Intelligent Generative Systems
Funding Source: Horizon Europe Framework Programme (2021-2027) under the grant agreement No 101215032.
Duration: 2025-2028 - Title: ResTick: Resilience Enhancement for Ticks and Tick-Borne Diseases in Sub-Saharan Africa
Funding Source: HORIZON JU Research and Innovation Actions (in collaboration with IMBB)
Duration: 2025-2028 - Title: FOR-SOLAR: Statistical Machine learning methods for output forecasting in grid connected and off-grid PV systems
Funding Source: SAUDI ARAMCO TECHNOLOGIES COMPANY
Duration: 2025-2026 - Title: NEMO-Tools: Next-generation monitoring and mapping tools to assess marine ecosystems and biodiversity
Funding Source: Hellenic Foundation for Research and Innovation
Duration: 2024-2026 - Title: Causal discovery and inference for surrogate-assisted optimization
Funding Source and funding scheme: Research and Development Agreement among Huawei Technologies (Ireland) C.O and FORTH
Duration: 2021-2027
B. COMPLETED PROJECTS
- Title: Disentangled representation learning via Mutual Information optimization with applications in speech representation learning
Funding Source: Private sector
Duration: 2024-2025 - Title: STOMA: Towards real-time, enhanced text-to-speech synthesis on the device
Funding Source: Hellenic Foundation for Research and Innovation
Duration: 2022-2025 - Title: FUSING: Biophysical tools FUSed via integrative computational approaches to decode protein foldING
Funding Source: FORTH Synergy
Duration: 2022-2024 - Title: SCALINCS: Scaling stochastic dynamics: from microscopic interactions to macroscopic phenomena
Funding Agency and funding scheme: Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “First Call for H.F.R.I. Research Projects to support faculty members and researchers and the procurement of high-cost research equipment”
Duration: 2020-2024 - Title: Data Landscaping: Traffic and Mobility Data Sources of Official Statistics
Funding Source: Eurostat
Duration: 2022 - Title: SOLAR-P: Evaluation of alternative solar panel technologies, computation of irradiance daily profiles
Funding source: Saudi Aramco and KAUST
Duration: 2021-2022 - Title: Characterising population dynamics with applications in biological data
Funding source: ESPA - Department of Development
Duration: 2020-2021 - Title: WNRG: Forecasting hourly wind-farm outputs based on wind-speed predictions from alternative providers
Funding source: EREN-Hellas
Duration: 2020 - Title: ENRICH: Enriched communication across the lifespan
Funding source: EU Horizon 2020, MSCA-ETN-2020
Duration: 2017-2020
PUBLICATIONS
-
2026
- T Aravanis, V Stojnić, B Psomas, N Komodakis, G Tolias (2026) Retrieve and Segment: Are a Few Examples Enough to Bridge the Supervision Gap in Open-Vocabulary Segmentation?, arXiv preprint arXiv:2602.23339
- E.H. Georgoulis, A. Papapantoleon, C. Smaragdakis (2026) A deep implicit-explicit minimizing movement method for partial integro-differential equations, with application to option pricing in jump-diffusion models, Communications in Nonlinear Science and Numerical Simulation, Vol. 156, 109709, https://doi.org/10.1016/j.cnsns.2026.109709
- 2025
- 2024
- 2023
- 2022
- 2021
- 2020
- 2019
PEOPLE
- Biza Konstantina (PhD candidate)
- Georgoulis Elias (MSc candidate)
- Kofidis Andreas (MSc candidate)
- Litsas Anastasios (MSc candidate)
- Papadaki Maria-Eleni (MSc candidate)
- Raptakis Michail (PhD candidate)
CONTACT US
Data Science Group,
Institute of Applied and Computational Mathematics,
Foundation for Research and Technology - Hellas
Nikolaou Plastira 100, Vassilika Vouton,
GR 700 13 Heraklion, Crete
GREECE
Tel: +30 2810 391800
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it. (Mrs. Maria Papadaki)
Tel.: +30 2810 391805
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it. (Mrs. Yiota Rigopoulou)