 Institute of Applied and Computational Mathematics
IACM is one of the few research institutes in Europe dedicated to promoting the use of advanced mathematics in natural sciences and engineering
Institute of Applied and Computational Mathematics
IACM is one of the few research institutes in Europe dedicated to promoting the use of advanced mathematics in natural sciences and engineering
Molecular Modelling
ABOUT
The
main focus of the Molecular Modelling group concerns the development of
mathematical and computational methodologies for molecular and biomolecular
materials. This involves several research areas including molecular simulations,
statistical mechanics, mathematical coarse-graining, stochastic processes and
data analysis for molecular models at equilibrium as well as under
non-equilibrium conditions. Our
group has extensive research experience in molecular dynamics simulations, in Monte
Carlo methods as well as in hierarchical multi-scale approaches combining
atomistic and coarse-grained models for complex molecular systems. We develop
novel computational methodologies for atomistic and coarse-grained models. We
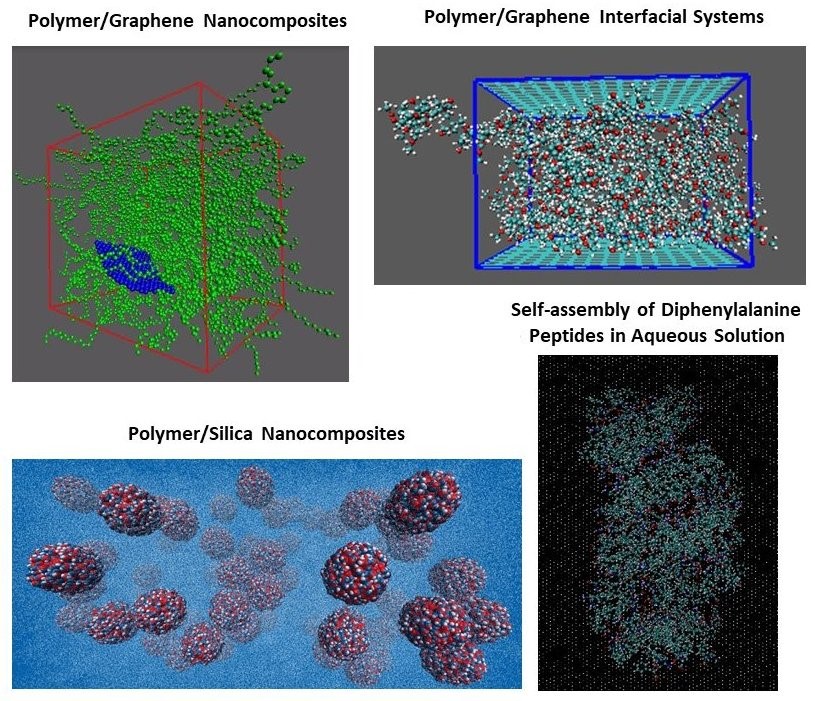
further apply such novel methods to a very broad range of systems/materials of
scientific and technological interest, such as nanocomposites, polymers,
graphene based nanostructured systems and biomolecules. Our
research group also develops and delivers methodological approaches and software
tools to address research projects/problems in all above areas.
RESEARCH AND DEVELOPMENT ACTIVITIES
The team is working on the development of novel mathematical and computational methods for the study of molecular systems/materials across multiple length and time scales, and in combining these methods with statistical analysis and data mining approaches.
More specifically the team is oriented in the following research directions:
- Development of mathematical methodologies for obtaining coarse-grained (CG) models for molecular systems at non-equilibrium conditions, e.g. under shear flow.
- Extension of variational inference path-space methods for obtaining CG models via data mining and machine learning methodologies and application of the new CG models in molecular systems at equilibrium and non-equilibrium conditions.
- Study of polymer nanocomposites via hierarchical multi-scale simulation approaches.
- Prediction of the properties of graphene sheets in graphene-based polymer nanocomposites.
- Study biomolecular systems (peptides, proteins) via molecular dynamics simulations and bioinformatics approaches.
It is a strategic objective of the group to extend existing synergies with other groups from outside Greece, as well as with research teams at FORTH, for combining different simulation methods, and/or simulations with experiment, in order to provide a fundamental understanding of materials behaviour.
Education and Training: The group contributes to the education and training of undergraduate, graduate and post-graduate students as well as of PhD candidates and Postdoctoral researchers in the area of Molecular Modelling.
Molecular Modelling
RESEARCH AND DEVELOPMENT PROGRAMS
A. ONGOING PROJECTS
- Title: Computational modeling of polymer solutions with metal nanoparticles,
Founding Agency: TOYOTA MOTOR CORPORATION
Duration: 2025-2026 - Title: ENGAGE: Enabling the Next-Generation of Computational Physicists and Engineers
Founding Agency: FORTH TRANSFER FUNDING TO CyL
Duration: 2022-2026 - Title: Ανάπτυξη μεθόδων Μοριακών Προσομοιώσεων πολυμερικών νανοσύνθετων υλικών
Founding Agency: THE GOODYEAR TIRE & RUBBER COMPANY
Duration: 2017-2027
B. COMPLETED PROJECTS
- Title: THUNDER, Thermal stimuli-responsive 3D printed electroactive polymer nanocomposites towards 4D “programmable” geometries,
Founding Agency: Hellenic Foundation for Research and Innovation (HFRI), Contract no. 15515
Duration: 2024-2025 - Title: MagMASim: Reconstructing the MAGnetic field of the Milky way via Astrophysical Techniques and Numerical SIMulations
Founding Agency: FORTH SYNERGY
Duration: 2020-2024
PUBLICATIONS
-
2026
- S Ektirici, V Harmandaris, CN Likos, TS Alexiou (2026) Ion-Modulated Polyelectrolyte Complexation of DNA and Polyacrylic Acid from Molecular Dynamics Simulations, arXiv preprint arXiv:2601.20541
- N Patsalidis, V Harmandaris (2026) The fascinating behavior of polymers at interfaces via molecular dynamics simulations, Computational Methods for the Multiscale Modeling of Soft Matter, 319-350, https://doi.org/10.1016/B978-0-44-327314-8.00018-X
- 2025
- 2024
- 2023
- 2022
- 2021
- 2020
- 2019
- 2018
- 2017
- 2016
- 2015
- 2014
- 2013
PEOPLE
RESEARCHERS
- Bačova, Petra
- Harmandaris, Vagelis
- Kalligiannaki, Evangelia
- Makridakis, Charalambos
- Rissanou, Anastassia
CONTACT US
For any information regarding the group please contact:
Molecular Modelling Group,
Institute of Applied and Computational Mathematics,
Foundation for Research and Technology - Hellas
Nikolaou Plastira 100, Vassilika Vouton,
GR 700 13 Heraklion, Crete
GREECE
Tel: +30 2810 391800
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it. (Mrs. Maria Papadaki)
Tel.: +30 2810 391805
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it. (Mrs. Yiota Rigopoulou)
Molecular Modelling Group,
Institute of Applied and Computational Mathematics,
Foundation for Research and Technology - Hellas
Nikolaou Plastira 100, Vassilika Vouton,
GR 700 13 Heraklion, Crete
GREECE
Tel: +30 2810 391800
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it. (Mrs. Maria Papadaki)
Tel.: +30 2810 391805
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it. (Mrs. Yiota Rigopoulou)